2.
df <- read.csv("http://goo.gl/Zjr9aF")
row.names(df) <- df$speaker
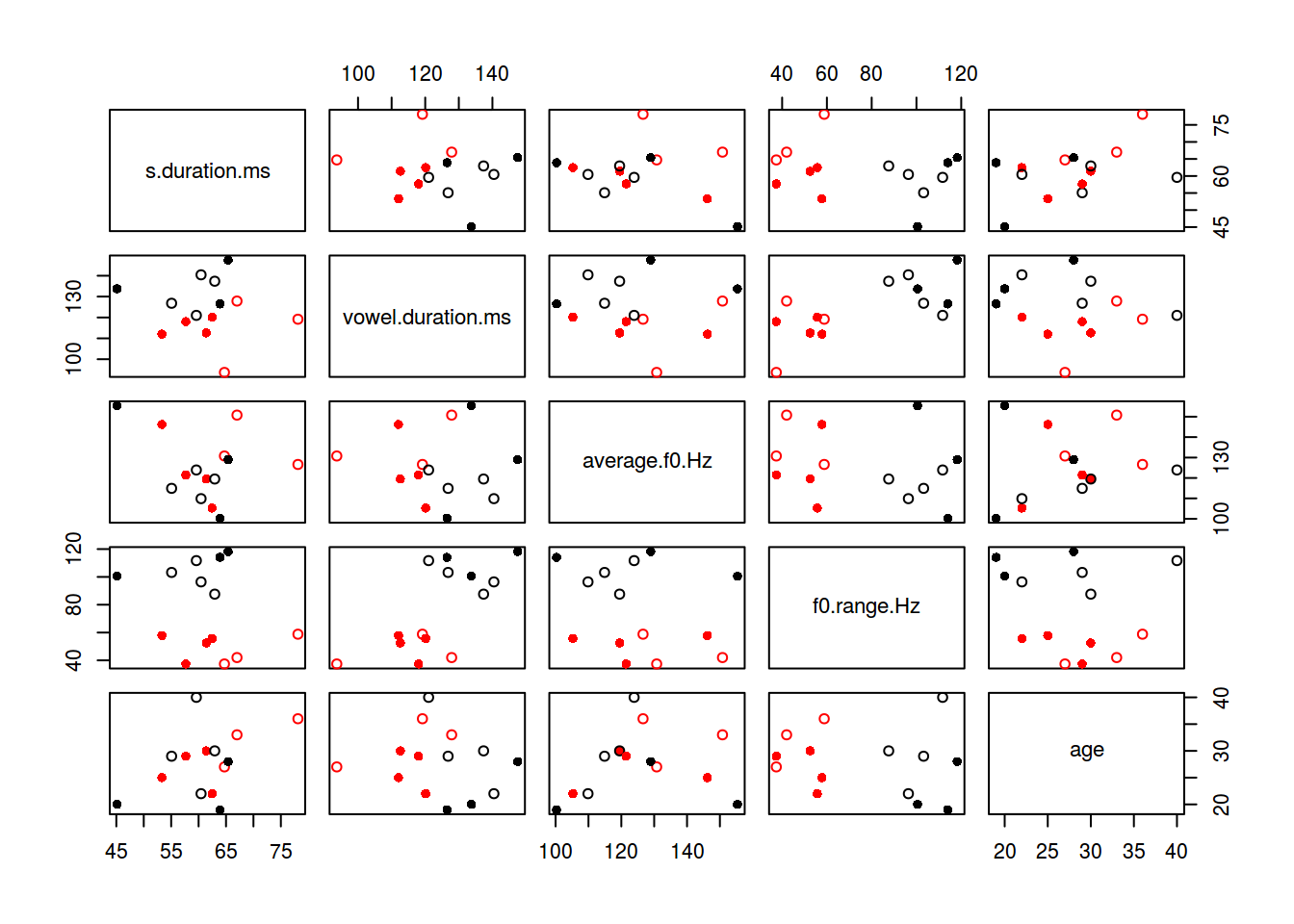
d <- dist((df[,-c(6:9, 1)]))
min(d)
## [1] 15.79903
set.seed(42)
km <- kmeans(d, 2)
cbind.data.frame(cluster = km$cluster, orientation = df$orientation) %>%
count(cluster, orientation)
plot(df[,-c(6:9, 1)], col = km$cluster, pch = c(16, 1)[df$orientation])
df %>%
mutate(cluster = km$cluster) %>%
group_by(cluster) %>%
summarise(mean = mean(perceived.as.homo.percent),
sd = sd(perceived.as.homo.percent))

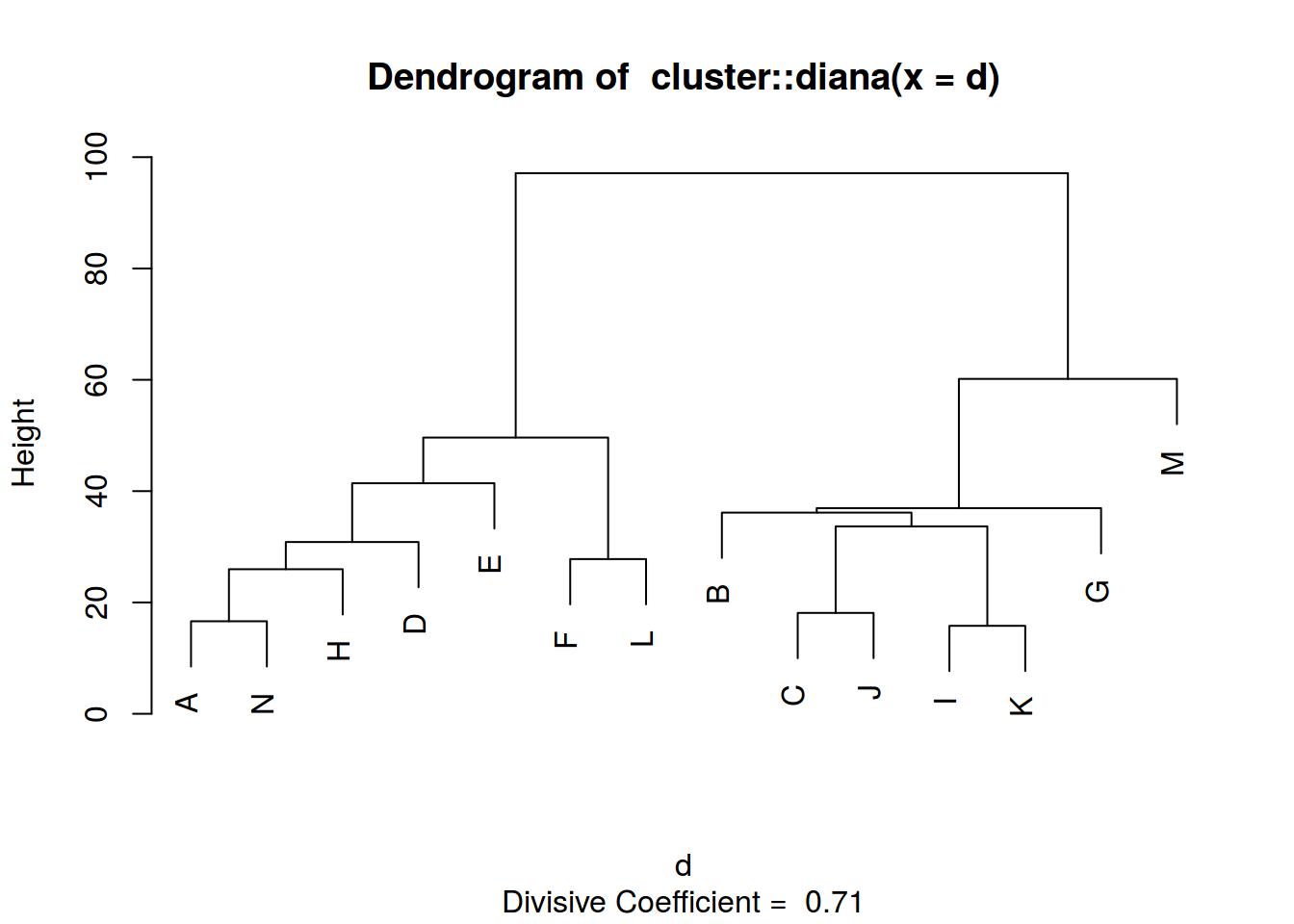
diana <- cluster::diana(d)
plot(diana)

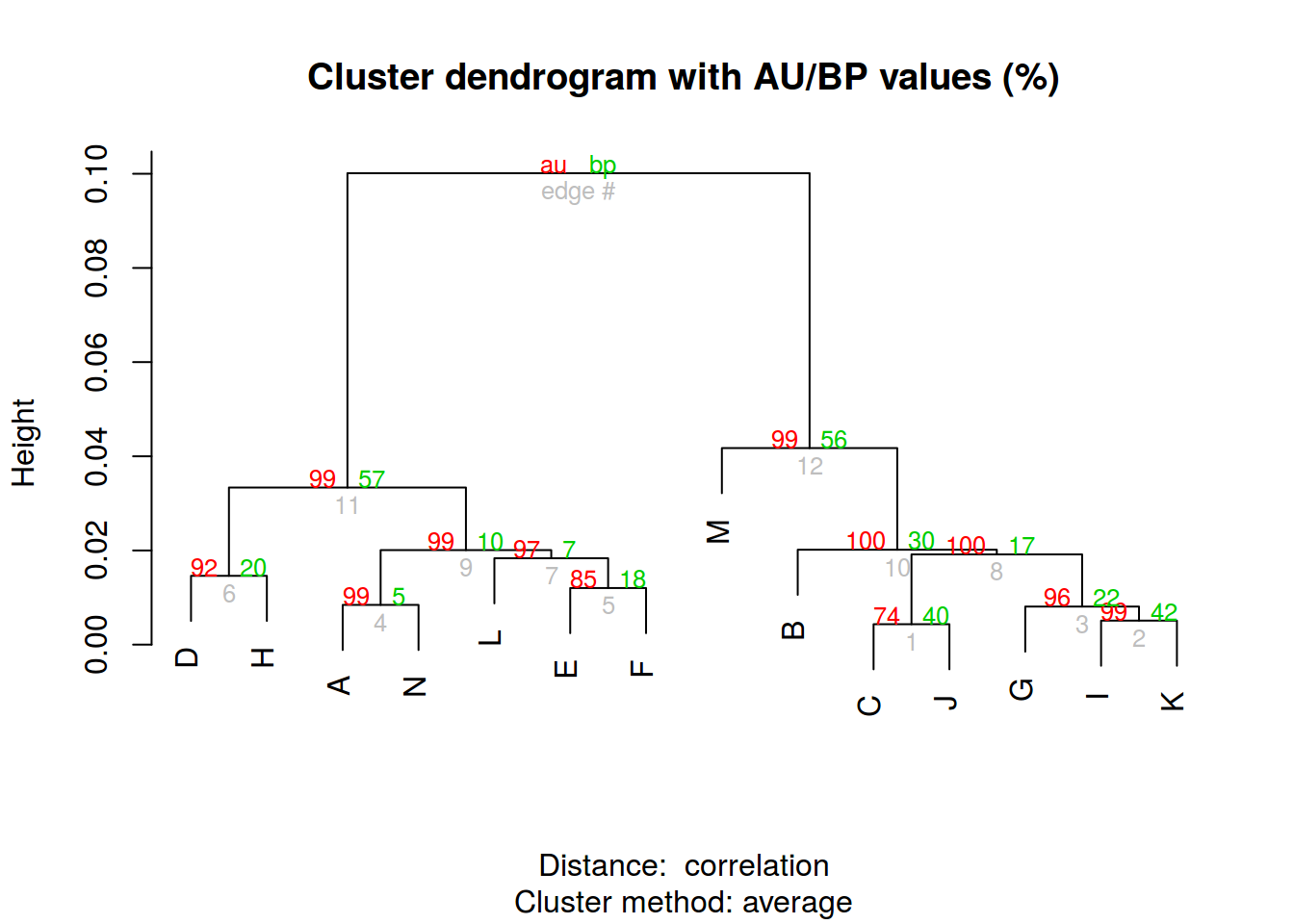
p <- pvclust::pvclust(t(df[,-c(1, 9, 10)]),
method.dist="cor",
method.hclust="average",
nboot=100)
## Bootstrap (r = 0.43)... Done.
## Warning: inappropriate distance matrices are omitted in computation: r =
## 0.428571428571429
## Bootstrap (r = 0.57)... Done.
## Bootstrap (r = 0.57)... Done.
## Bootstrap (r = 0.71)... Done.
## Bootstrap (r = 0.86)... Done.
## Bootstrap (r = 1.0)... Done.
## Bootstrap (r = 1.0)... Done.
## Bootstrap (r = 1.14)... Done.
## Bootstrap (r = 1.29)... Done.
## Bootstrap (r = 1.29)... Done.
## Warning in a$p[] <- c(1, bp[r == 1]): number of items to replace is not a
## multiple of replacement length
3
library(ape)
df <- read.csv("https://goo.gl/4sJqv1")
data <- df[,-c(1:3)]
row.names(data) <- paste(1:294, df$english)
data %>%
dist() %>%
hclust() ->
hc
plot(as.phylo(hc),
type = "fan",
cex = 0.6,
no.margin = TRUE,
tip.color = c("red",
"navy")[as.factor(df$archi)])
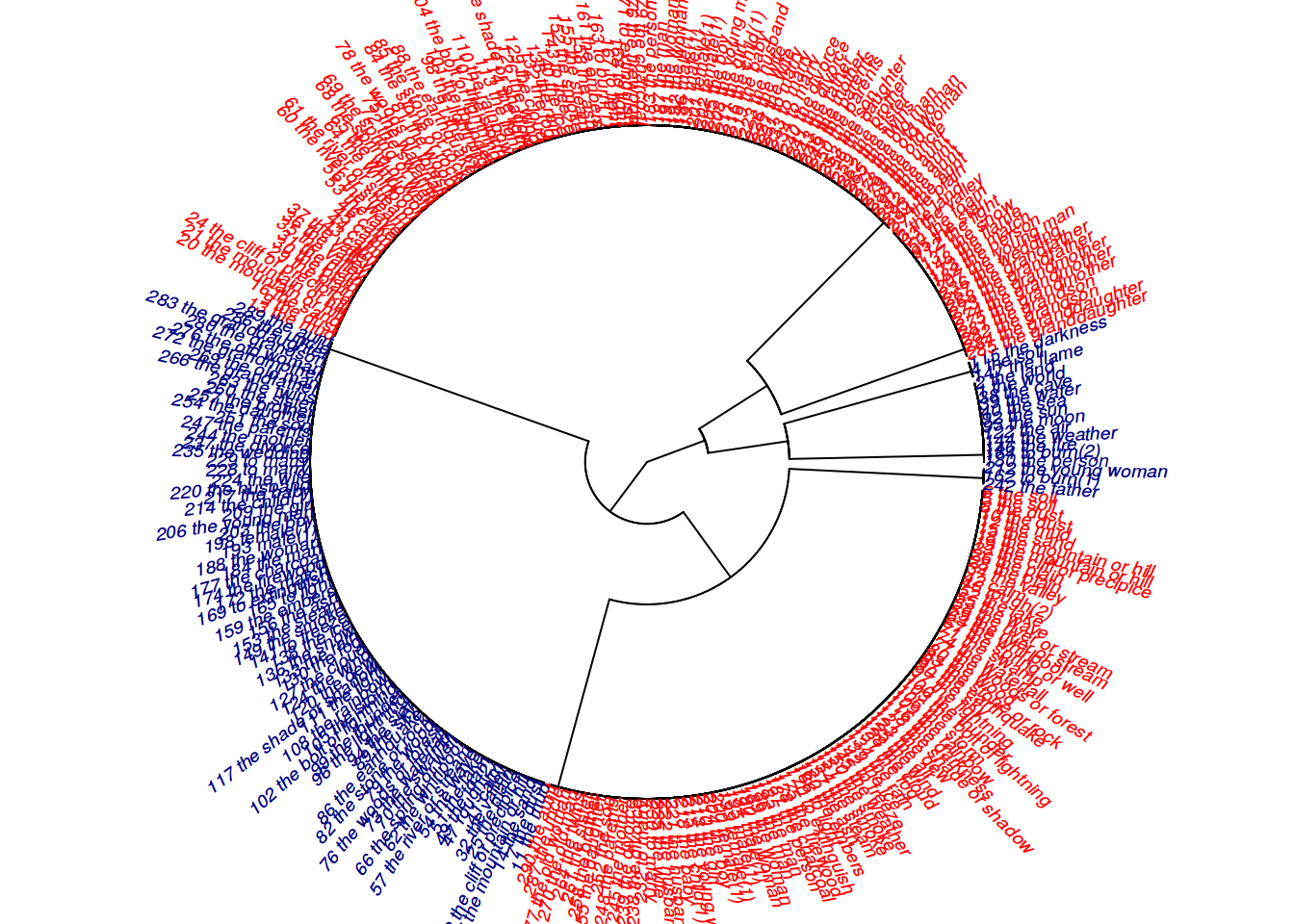
- blue are the Bezhta unique words
- red are the Archi unique words









