1. Consonants and vowels
lapsyd <- read.csv("https://goo.gl/eD4S5n")
map.feature(lapsyd$name, features = lapsyd$area,
label = lapsyd$name,
label.hide = TRUE)
## Warning: There is no coordinates for languages Gelao, Naxi, Nepali
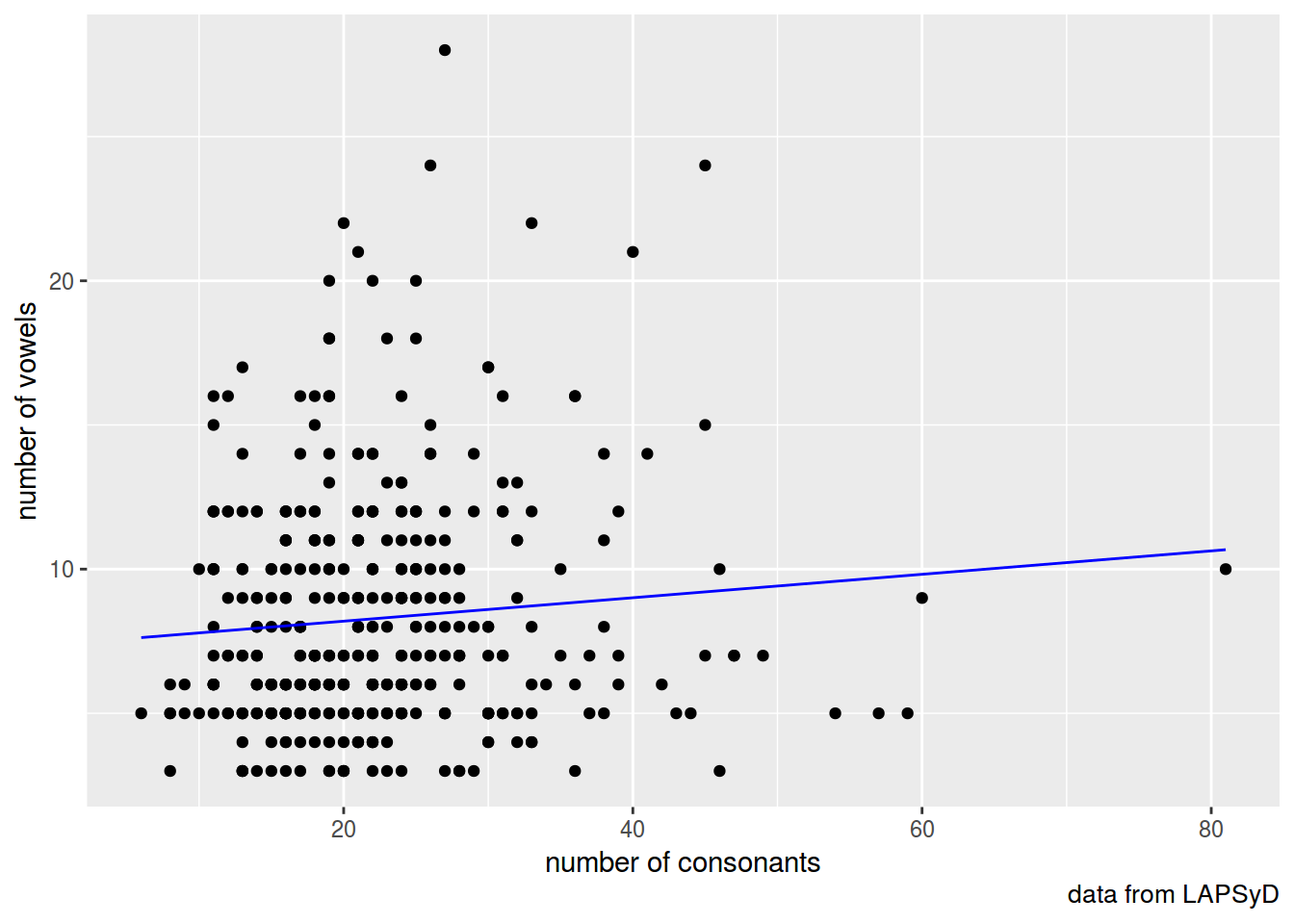
fit1 <- lm(count_vowel~count_consonant, data = lapsyd)
summary(fit1)
##
## Call:
## lm(formula = count_vowel ~ count_consonant, data = lapsyd)
##
## Residuals:
## Min 1Q Median 3Q Max
## -6.252 -2.992 -1.195 2.201 19.520
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 7.38236 0.54784 13.475 <2e-16 ***
## count_consonant 0.04065 0.02274 1.787 0.0746 .
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 4.093 on 400 degrees of freedom
## Multiple R-squared: 0.007923, Adjusted R-squared: 0.005443
## F-statistic: 3.195 on 1 and 400 DF, p-value: 0.07464
lapsyd$model1 <- predict(fit1)
lapsyd %>%
ggplot(aes(count_consonant, count_vowel))+
geom_point()+
geom_line(aes(count_consonant, model1), color = "blue")+
labs(x = "number of consonants",
y = "number of vowels",
caption = "data from LAPSyD")
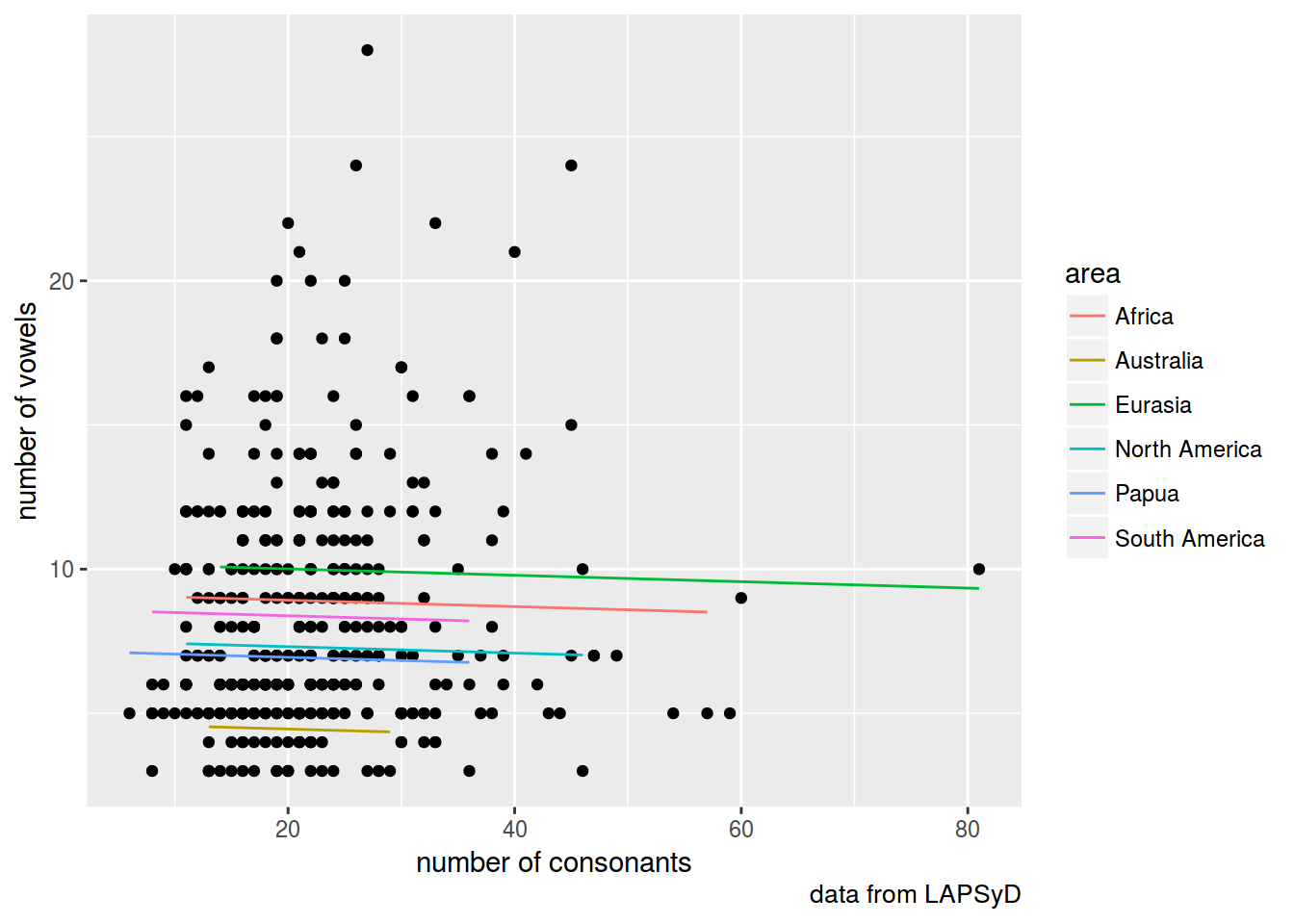
fit2 <- lmer(count_vowel ~ count_consonant + (1|area), data = lapsyd)
summary(fit2)
## Linear mixed model fit by REML ['lmerMod']
## Formula: count_vowel ~ count_consonant + (1 | area)
## Data: lapsyd
##
## REML criterion at convergence: 2242
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -1.5298 -0.6147 -0.2599 0.5146 4.7043
##
## Random effects:
## Groups Name Variance Std.Dev.
## area (Intercept) 3.983 1.996
## Residual 14.753 3.841
## Number of obs: 402, groups: area, 6
##
## Fixed effects:
## Estimate Std. Error t value
## (Intercept) 7.8928 0.9852 8.011
## count_consonant -0.0111 0.0240 -0.463
##
## Correlation of Fixed Effects:
## (Intr)
## cont_cnsnnt -0.518
lapsyd$model2 <- predict(fit2)
lapsyd %>%
ggplot(aes(count_consonant, count_vowel))+
geom_point()+
geom_line(aes(count_consonant, model2, color = area))+
labs(x = "number of consonants",
y = "number of vowels",
caption = "data from LAPSyD")
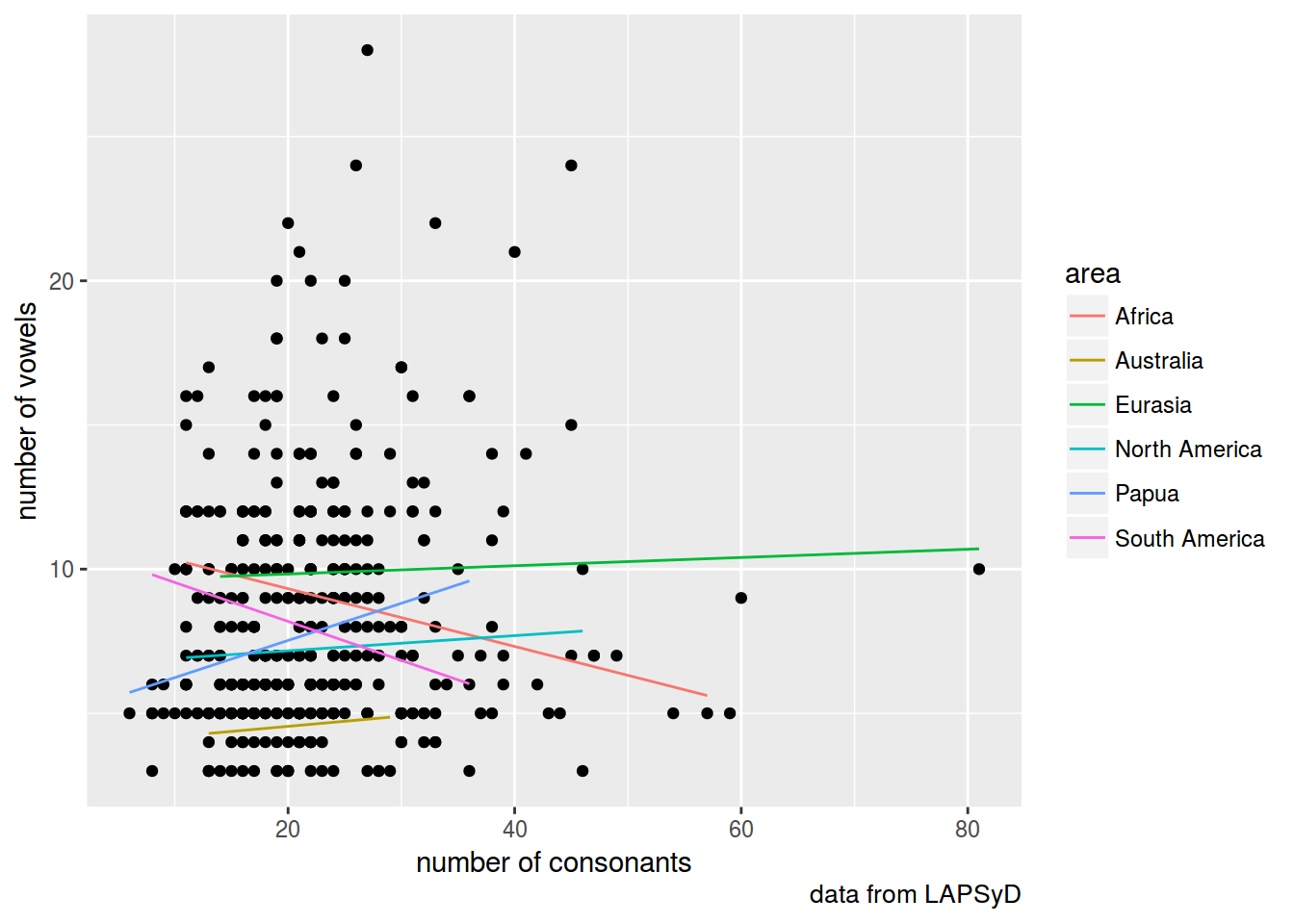
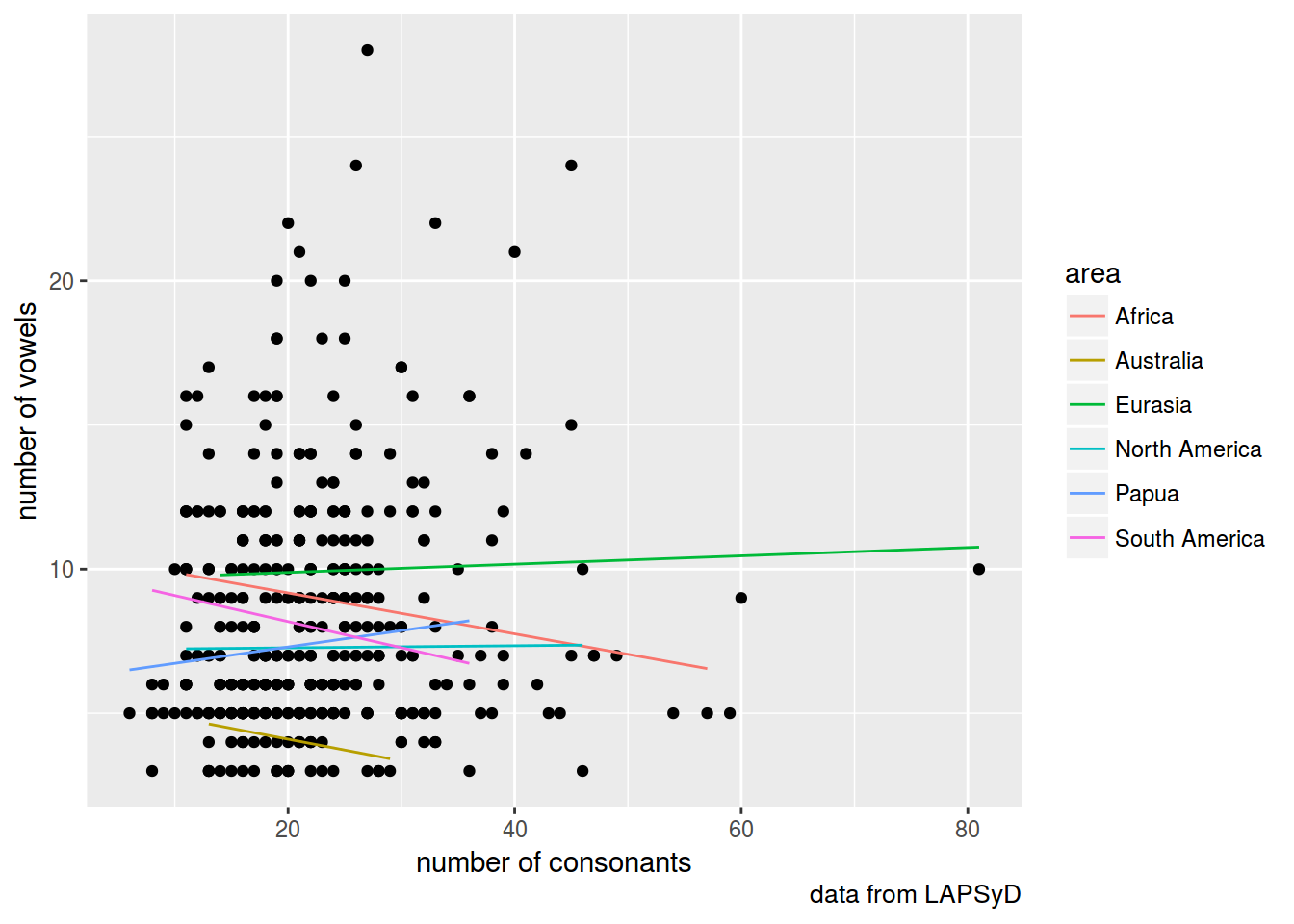
fit3 <- lmer(count_vowel ~ count_consonant + (1+count_consonant|area), data = lapsyd)
summary(fit3)
## Linear mixed model fit by REML ['lmerMod']
## Formula: count_vowel ~ count_consonant + (1 + count_consonant | area)
## Data: lapsyd
##
## REML criterion at convergence: 2236.4
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -1.7991 -0.6217 -0.2334 0.5299 4.7743
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## area (Intercept) 11.94558 3.4562
## count_consonant 0.01355 0.1164 -0.84
## Residual 14.32212 3.7845
## Number of obs: 402, groups: area, 6
##
## Fixed effects:
## Estimate Std. Error t value
## (Intercept) 7.866281 1.555096 5.058
## count_consonant -0.005108 0.056987 -0.090
##
## Correlation of Fixed Effects:
## (Intr)
## cont_cnsnnt -0.854
lapsyd$model3 <- predict(fit3)
lapsyd %>%
ggplot(aes(count_consonant, count_vowel))+
geom_point()+
geom_line(aes(count_consonant, model3, color = area))+
labs(x = "number of consonants",
y = "number of vowels",
caption = "data from LAPSyD")
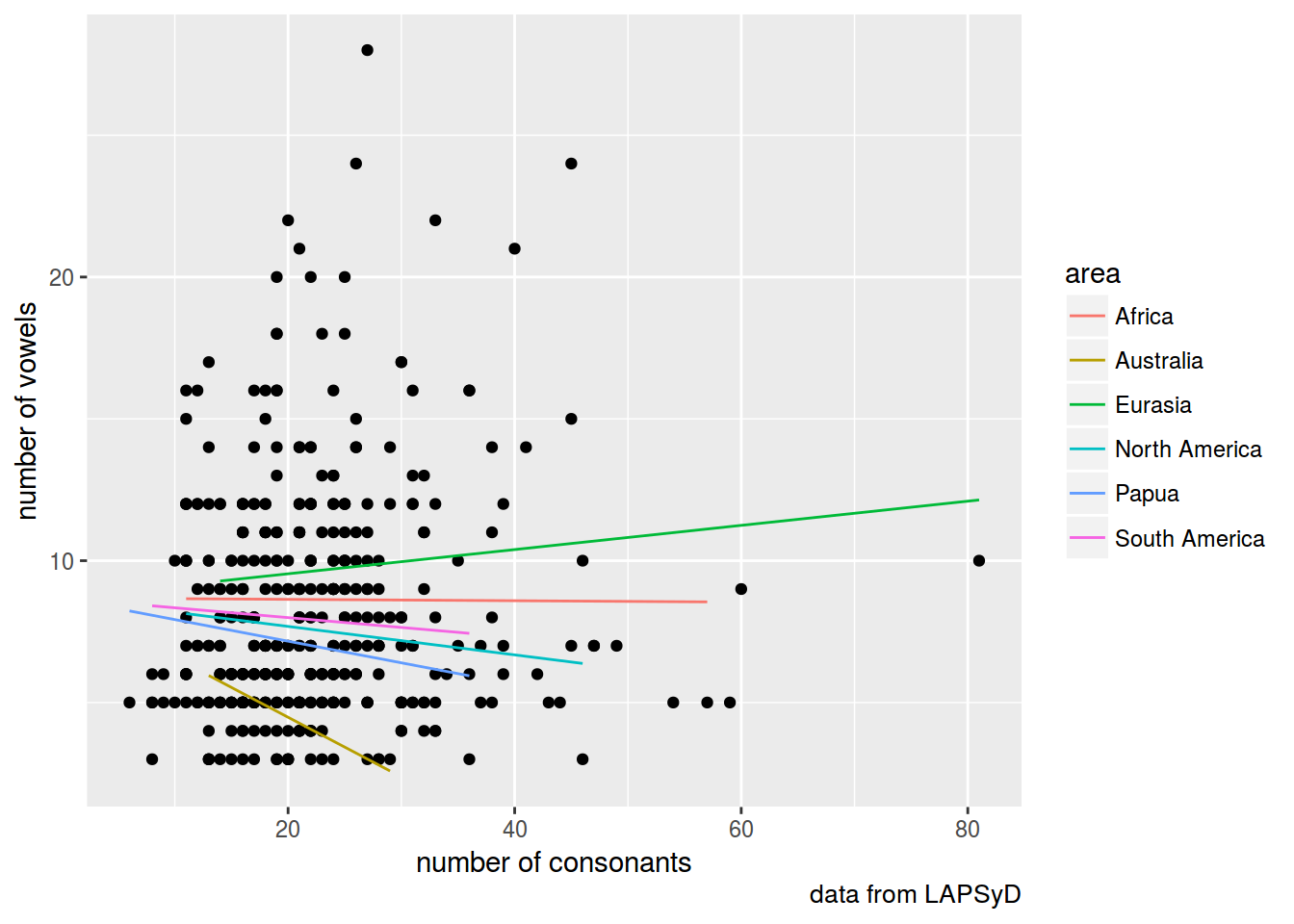
fit4 <- lmer(count_vowel ~ count_consonant + (0+count_consonant|area), data = lapsyd)
summary(fit4)
## Linear mixed model fit by REML ['lmerMod']
## Formula: count_vowel ~ count_consonant + (0 + count_consonant | area)
## Data: lapsyd
##
## REML criterion at convergence: 2250.8
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -1.5967 -0.6774 -0.2688 0.4497 4.6750
##
## Random effects:
## Groups Name Variance Std.Dev.
## area count_consonant 0.008151 0.09028
## Residual 15.093094 3.88498
## Number of obs: 402, groups: area, 6
##
## Fixed effects:
## Estimate Std. Error t value
## (Intercept) 8.68579 0.59439 14.613
## count_consonant -0.05519 0.04649 -1.187
##
## Correlation of Fixed Effects:
## (Intr)
## cont_cnsnnt -0.563
lapsyd$model4 <- predict(fit4)
lapsyd %>%
ggplot(aes(count_consonant, count_vowel))+
geom_point()+
geom_line(aes(count_consonant, model4, color = area))+
labs(x = "number of consonants",
y = "number of vowels",
caption = "data from LAPSyD")
fit5 <- lmer(count_vowel ~ count_consonant + (1|area) + (0+count_consonant|area), data = lapsyd)
summary(fit5)
## Linear mixed model fit by REML ['lmerMod']
## Formula:
## count_vowel ~ count_consonant + (1 | area) + (0 + count_consonant |
## area)
## Data: lapsyd
##
## REML criterion at convergence: 2239.8
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -1.6499 -0.6023 -0.2494 0.5018 4.7428
##
## Random effects:
## Groups Name Variance Std.Dev.
## area (Intercept) 5.67146 2.38148
## area.1 count_consonant 0.00606 0.07785
## Residual 14.42645 3.79822
## Number of obs: 402, groups: area, 6
##
## Fixed effects:
## Estimate Std. Error t value
## (Intercept) 8.19243 1.15107 7.117
## count_consonant -0.02700 0.04326 -0.624
##
## Correlation of Fixed Effects:
## (Intr)
## cont_cnsnnt -0.337
lapsyd$model5 <- predict(fit5)
lapsyd %>%
ggplot(aes(count_consonant, count_vowel))+
geom_point()+
geom_line(aes(count_consonant, model5, color = area))+
labs(x = "number of consonants",
y = "number of vowels",
caption = "data from LAPSyD")
anova(fit5, fit4, fit3, fit2, fit1)
## refitting model(s) with ML (instead of REML)
## Data: lapsyd
## Models:
## fit1: count_vowel ~ count_consonant
## fit4: count_vowel ~ count_consonant + (0 + count_consonant | area)
## fit2: count_vowel ~ count_consonant + (1 | area)
## fit5: count_vowel ~ count_consonant + (1 | area) + (0 + count_consonant |
## fit5: area)
## fit3: count_vowel ~ count_consonant + (1 + count_consonant | area)
## Df AIC BIC logLik deviance Chisq Chi Df Pr(>Chisq)
## fit1 3 2277.8 2289.8 -1135.9 2271.8
## fit4 4 2254.8 2270.7 -1123.4 2246.8 25.0256 1 5.657e-07 ***
## fit2 4 2245.8 2261.8 -1118.9 2237.8 8.9793 0 < 2.2e-16 ***
## fit5 5 2246.9 2266.9 -1118.4 2236.9 0.9009 1 0.34254
## fit3 6 2245.7 2269.6 -1116.8 2233.7 3.2150 1 0.07297 .
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1




