1.1
df <- read.csv("http://goo.gl/Qo3Yy2")
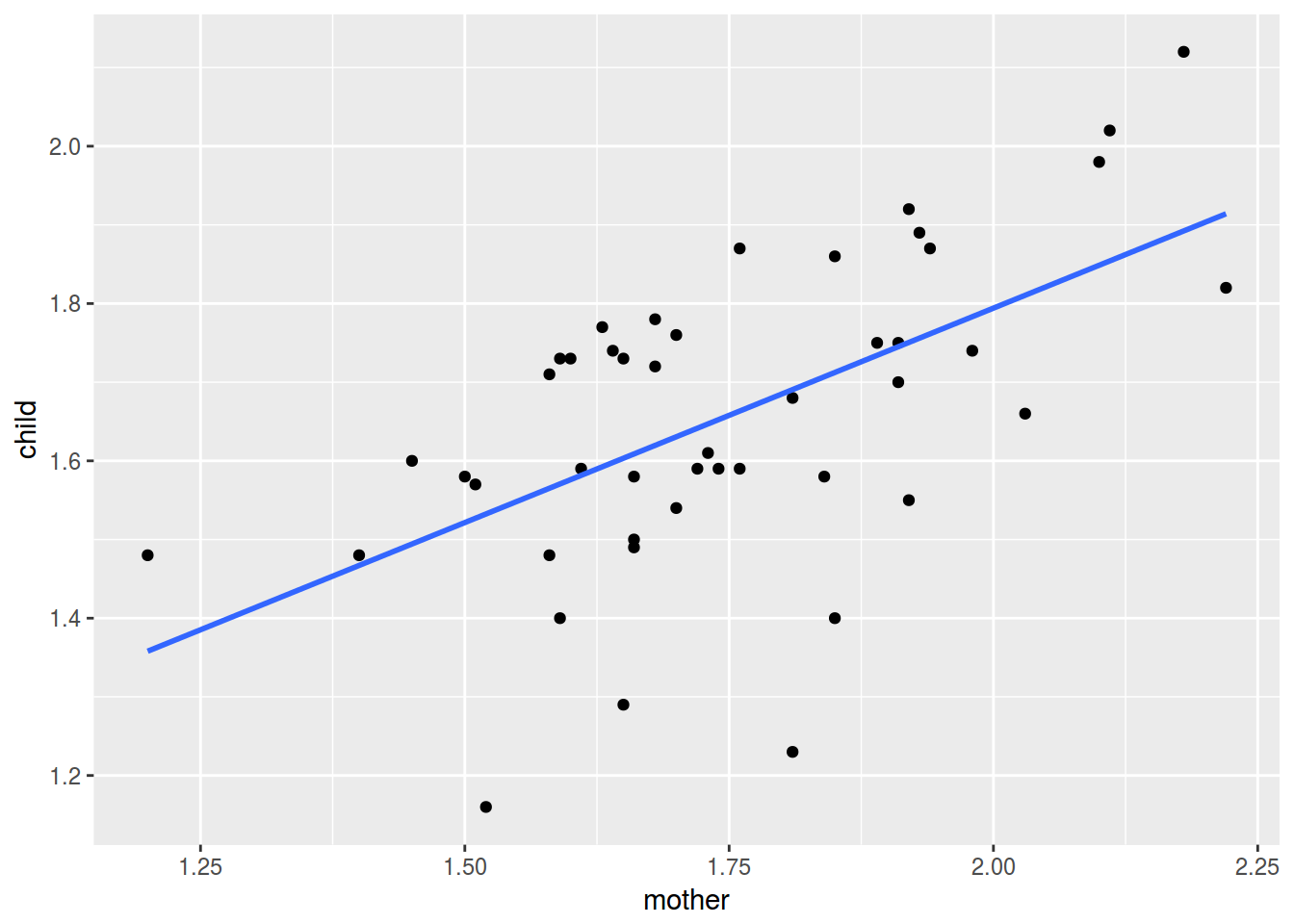
df %>%
ggplot(aes(mother, child))+
geom_point()+
geom_smooth(method = "lm", se = FALSE)
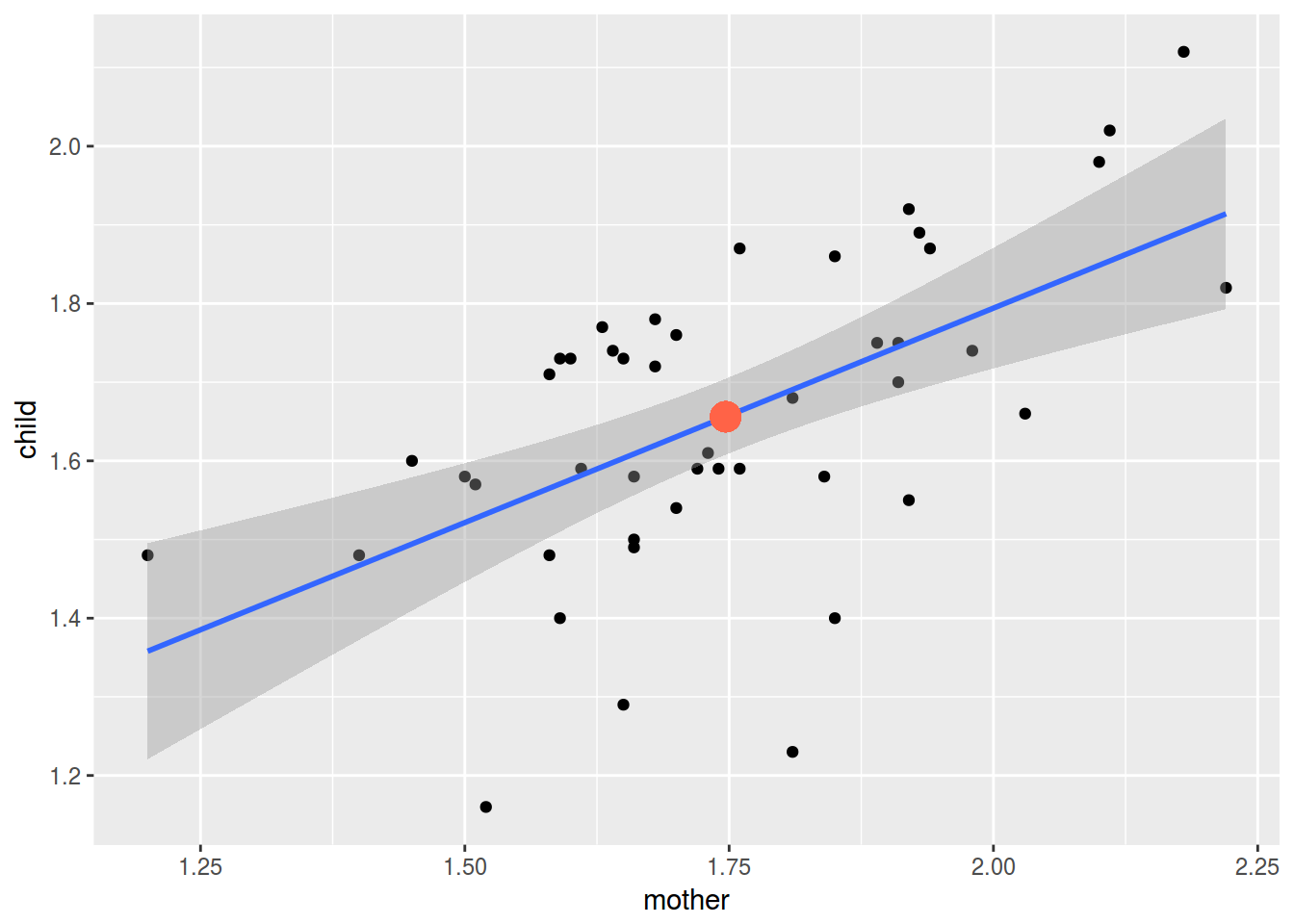
df %>%
ggplot(aes(mother, child))+
geom_point()+
geom_smooth(method = "lm")+
geom_point(aes(mean(mother), mean(child)), color = "tomato", size = 5)
cor(df)
## child mother
## child 1.0000000 0.5761599
## mother 0.5761599 1.0000000
fit <- lm(child~mother, data = df)
summary(fit)
##
## Call:
## lm(formula = child ~ mother, data = df)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.46058 -0.08925 0.01071 0.13333 0.22770
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.7038 0.2051 3.432 0.00132 **
## mother 0.5452 0.1166 4.676 2.79e-05 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.1627 on 44 degrees of freedom
## Multiple R-squared: 0.332, Adjusted R-squared: 0.3168
## F-statistic: 21.86 on 1 and 44 DF, p-value: 2.789e-05
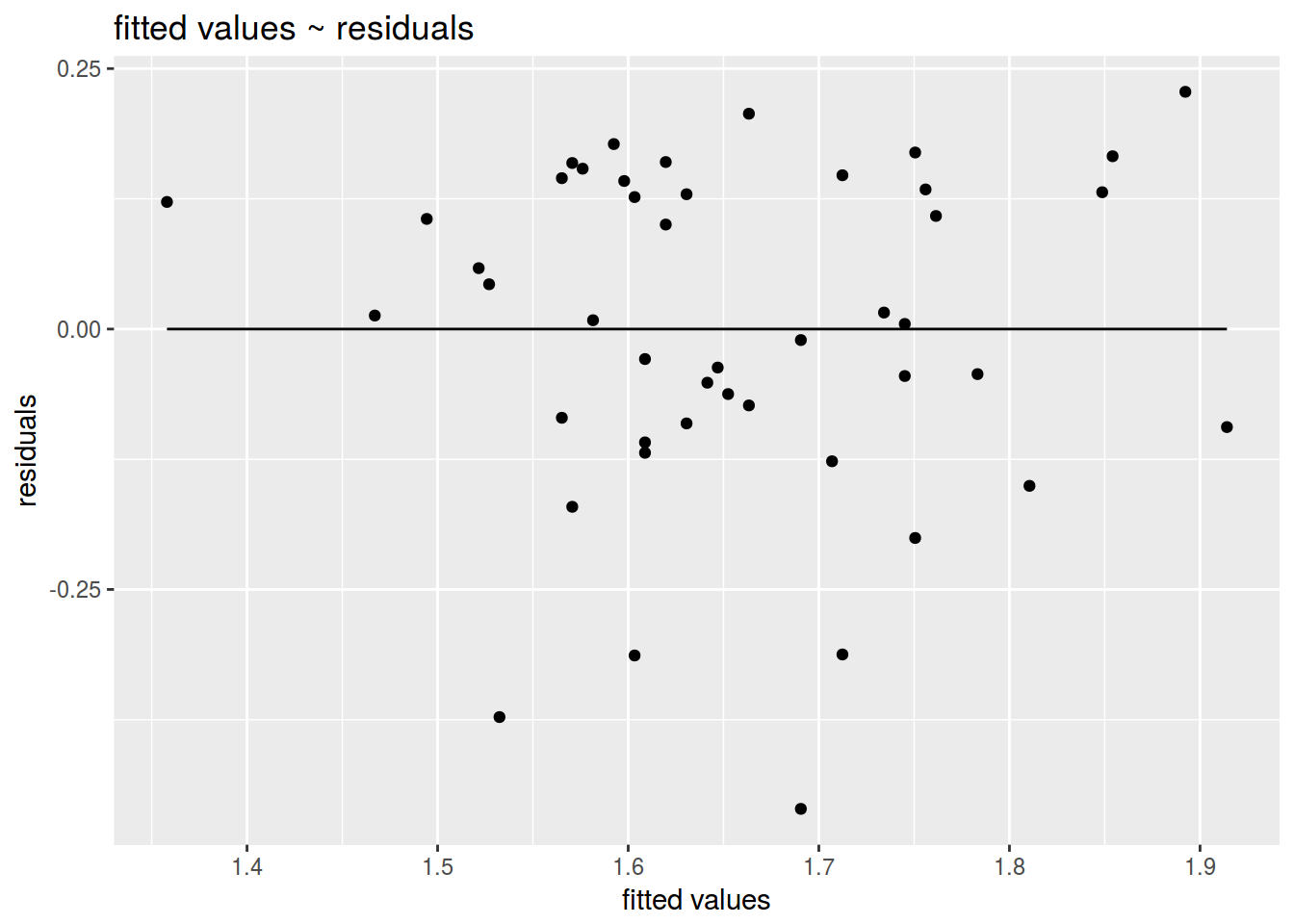
ggplot(data = fit, aes(fit$fitted.values, fit$residuals))+
geom_point()+
geom_line(aes(y = 0))+
labs(title = "fitted values ~ residuals",
x = "fitted values",
y = "residuals")
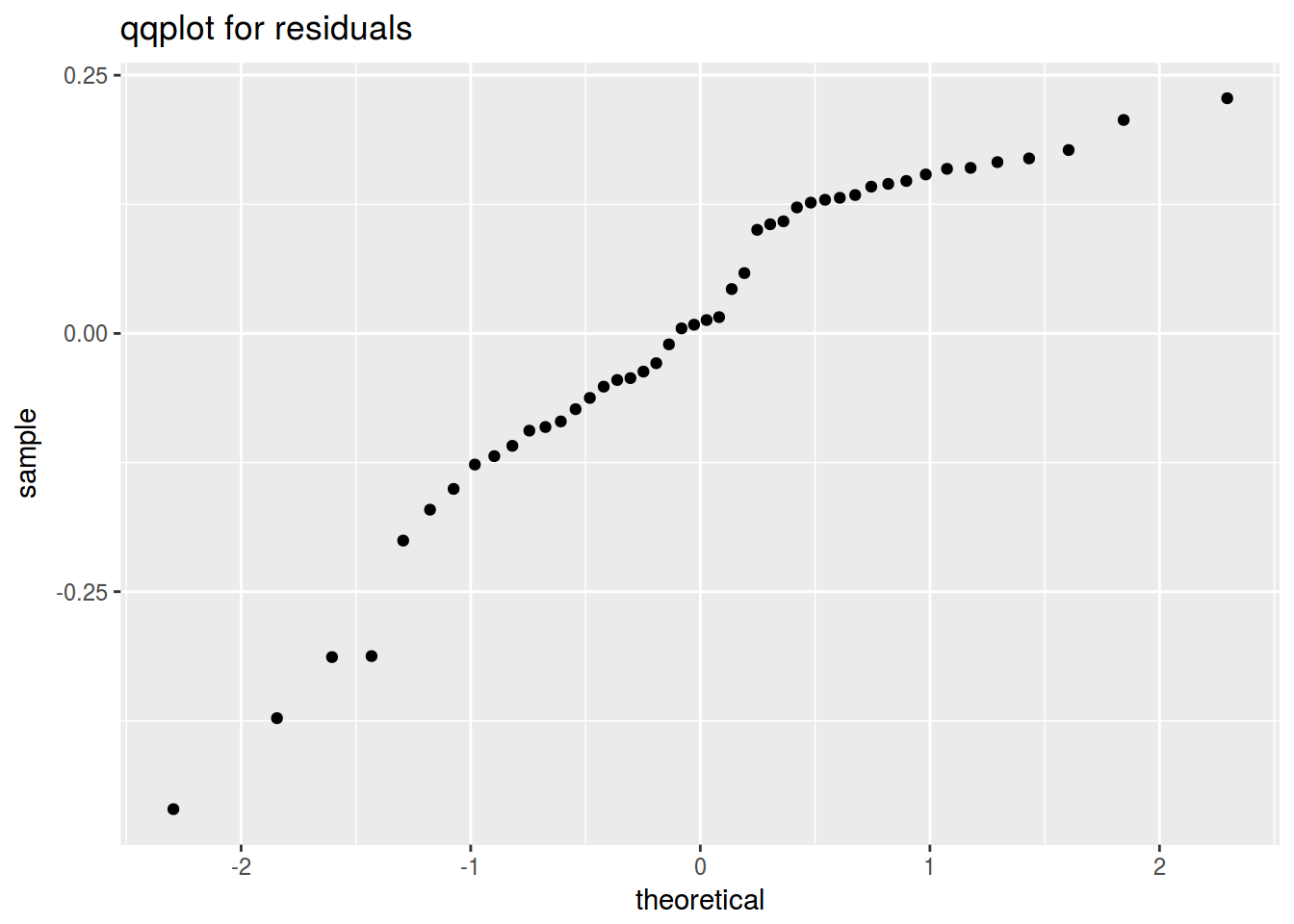
ggplot(data = fit, aes(sample = fit$residuals))+
geom_qq()+
labs(title = "qqplot for residuals")
predict(fit, data.frame(mother = 11:17/10))
## 1 2 3 4 5 6 7
## 1.303490 1.358009 1.412528 1.467048 1.521567 1.576086 1.630605
2.
df <- read.csv("https://goo.gl/TcyiRc", sep = "\t")
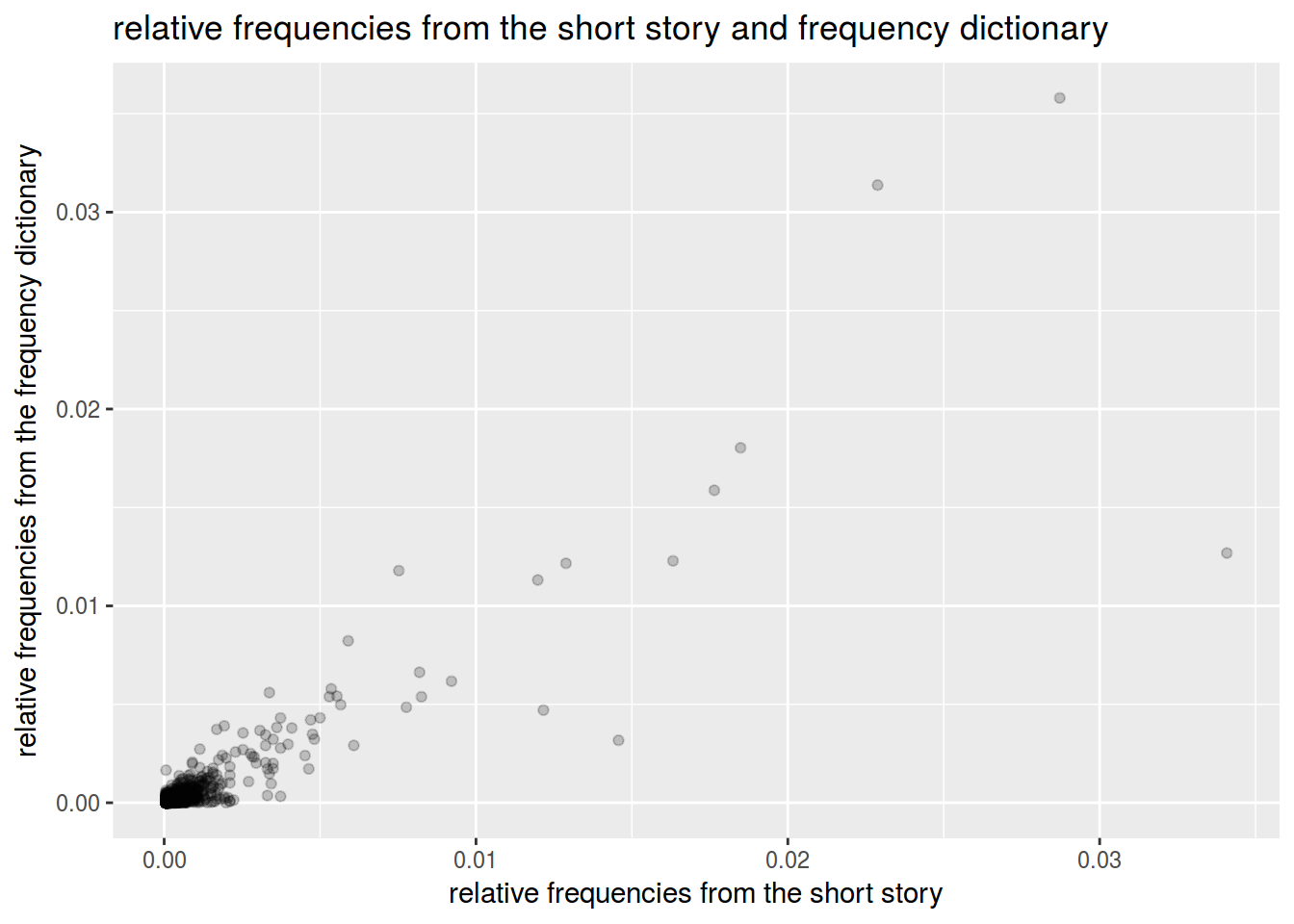
df %>%
ggplot(aes(r.frequency,rus.freq.dict))+
geom_point(alpha = 0.2)+
labs(titles = "relative frequencies from the short story and frequency dictionary",
x = "relative frequencies from the short story",
y = "relative frequencies from the frequency dictionary")
library(scales)
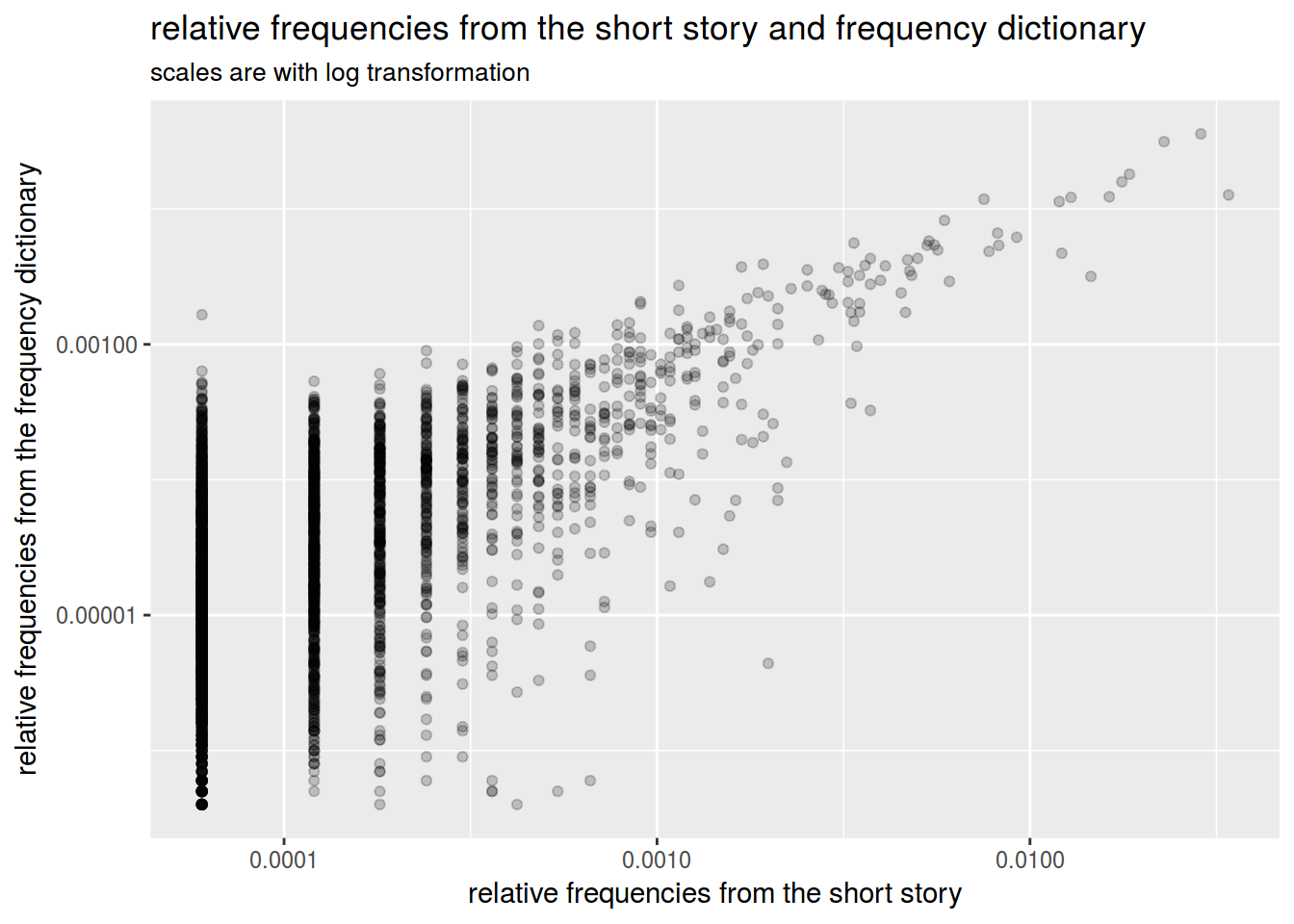
df %>%
ggplot(aes(r.frequency,rus.freq.dict))+
geom_point(alpha = 0.2)+
labs(titles = "relative frequencies from the short story and frequency dictionary",
subtitle = "scales are with log transformation",
x = "relative frequencies from the short story",
y = "relative frequencies from the frequency dictionary")+
scale_x_log10(labels = comma)+
scale_y_log10(labels = comma)